Implementation of a Non-Invasive Bioprospecting Protocol for Isolation of Lactobacillus from Feces of Hens Under Foraging Conditions
Main Article Content
Keywords
Bioprospecting, probiotics, lactic acid bacteria (LAB), antagonism test
Abstract
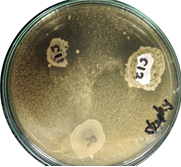
In animal production, probiotics seek to replace the use of antibiotics, while diminishing mortality and morbidity rates to raise productivity. Probiotics constitute a natural alternative that, in contrast with antibiotics, neither produces pathogen resistance, nor leaves chemical residues in the final product. Several bacteria, including some belonging to the genus Lactobacillus have been described as probiotics with high potential. A non-invasive bioprospecting protocol aimed for the isolation and characterization of lactobacilli from chicken feces was established. Fecal samples were collected from the ground. These were diluted and cultured in LAB selective medium. Colonies were identified by three methods: Gram stain, MALDI-TOF MS and sequencing of 16S rRNA gene. An initial probiotic potential of lactobacilli isolates was determined via antagonism tests using five enteropathogen reference strains: Staphylococcus aureus, Enterococcus faecium, Candida albicans, Pseudomonas spp. and Salmonella spp. 24 isolates belonging to four Lactobacillus species were identified by MALDITOF MS. BLAST of 16S rRNA gene of eight randomly selected isolates, confirmed MALDI-TOF MS identification. Five of these eight isolates inhibited the growth of at least one of the pathogenic strains used, three isolates of Lactobacillus plantarum and two of Lactobacillus salivarius. Our protocol achieved 21 lactobacilli per 100 isolates performance, greatly surpassing the normal percentage of lactobacilli in chicken gut microbiome, that so, its implementation would facilitate the isolation and identification of new probiotic strains from feces.
Downloads
References
H. C. Wegener, “Antibiotics in animal feed and their role in resistance development,” Current opinion in microbiology, vol. 6, no. 5, pp. 439–445, 2003. [Online]. Available: https://doi.org/10.1016/j.mib.2003.09.009
F. M. Aarestrup, “Occurrence of glycopeptide resistance among enterococcus faecium isolates from conventional and ecological poultry farms,” Microbial Drug Resistance, vol. 1, no. 3, pp. 255–257, 1995. [Online]. Available: https://doi.org/10.1089/mdr.1995.1.255
J. Bates, J. Z. Jordens, and D. T. Griffiths, “Farm animals as a putative reservoir for vancomycin-resistant enterococcal infection in man,” Journal of Antimicrobial Chemotherapy, vol. 34, no. 4, pp. 507–514, 1994. [Online]. Available: https://doi.org/10.1093/jac/34.4.507
I. Klare, H. Heier, H. Claus, G. Böhme, S. Marin, G. Seltmann, R. Hakenbeck, V. Antanassova, and W. Witte, “Enterococcus faecium strains with vana-mediated high-level glycopeptide resistance isolated from animal foodstuffs and fecal samples of humans in the community,” Microbial Drug Resistance, vol. 1, no. 3, pp. 265–272, 1995.
J. Stephenson, “Antibiotics in animal feed,” JAMA, vol. 290, no. 11, pp. 1443–1443, 2003.
F. Guarner and G. Schaafsma, “Probiotics.” International journal of food microbiology, vol. 39, no. 3, pp. 237–238, 1998.
F. Chaucheyras-Durand and H. Durand, “Probiotics in animal nutrition and health,” Beneficial microbes, vol. 1, no. 1, pp. 3–9, 2009. [Online]. Available: https://doi.org/10.3920/BM2008.1002
M. Vanbelle, E. Teller, and M. Focant, “Probiotics in animal nutrition: a review,” Archives of Animal Nutrition, vol. 40, no. 7, pp. 543–567, 1990. [Online]. Available: https://doi.org/10.1080/17450399009428406
P. Banjeree and N. Pradhan, “Live yeasts a good alternative to agp in broiler chickens,” World Poultry, vol. 22, no. 8, pp. 32–34, 2006. [Online]. Available: https://www.poultryworld.net/PageFiles/27778/001_ boerderij-download-WP6926D01.pdf
J. Higgins, S. Higgins, J. Vicente, A. Wolfenden, G. Tellez, and B. Hargis, “Temporal effects of lactic acid bacteria probiotic culture on salmonella in neonatal broilers,” Poultry Science, vol. 86, no. 8, pp. 1662–1666, 2007. [Online]. Available: https://doi.org/10.1093/ps/86.8.1662
S. Higgins, J. Higgins, A. Wolfenden, S. Henderson, A. Torres-Rodriguez, G. Tellez, and B. Hargis, “Evaluation of a lactobacillus-based probiotic culture for the reduction of salmonella enteritidis in neonatal broiler chicks,” Poultry Science, vol. 87, no. 1, pp. 27–31, 2008. [Online]. Available: https://doi.org/10.3382/ps.2007-00210
R. La Ragione, A. Narbad, M. Gasson, and M. J. Woodward, “In vivo characterization of lactobacillus johnsonii fi9785 for use as a defined competitive exclusion agent against bacterial pathogens in poultry,” Letters in Applied Microbiology, vol. 38, no. 3, pp. 197–205, 2004.
R. M. La Ragione and M. J. Woodward, “Competitive exclusion by bacillus subtilis spores of salmonella enterica serotype enteritidis and clostridium perfringens in young chickens,” Veterinary microbiology, vol. 94, no. 3, pp. 245–256, 2003. [Online]. Available: https://doi.org/10.1016/S0378-1135(03) 00077-4
G. Tellez, C. Pixley, R. Wolfenden, S. Layton, and B. Hargis, “Probiotics/direct fed microbials for salmonella control in poultry,” Food Research International, vol. 45, no. 2, pp. 628–633, 2012. [Online]. Available: https://doi.org/10.1016/j.foodres.2011.03.047
F. Yan, W. Wang, R. Wolfenden, and H. Cheng, “The effect of bacillus subtilis based probiotic on bone health in broiler chickens,” Poultry Science, vol. 95, p. 40, 2016. [Online]. Available: https://doi.org/10.1093/jas/sky092
T. Inatomi, “Growth performance, gut mucosal immunity and carcass and intramuscular fat of broilers fed diet containing a combination of three probiotics,” Sci Postprint, vol. 1, p. e00052, 2015.
V. Kurtoglu*, F. Kurtoglu, E. Seker, B. Coskun, T. Balevi, and E. Polat, “Effect of probiotic supplementation on laying hen diets on yield performance and serum and egg yolk cholesterol,” Food additives and contaminants, vol. 21, no. 9, pp. 817–823, 2004. [Online]. Available: https://doi.org/10.1080/02652030310001639530
C. Pineda-Quiroga, R. Atxaerandio, I. Zubiria, I. Gonzalez-Pozuelo, A. Hurtado, R. Ruiz, and A. Garcia-Rodriguez, “Productive performance and cecal microbial counts of floor housed laying hens supplemented with dry whey powder alone or combined with pediococcus acidilactici in the late phase of production,” Livestock Science, vol. 195, pp. 9–12, 2017. [Online]. Available: https://doi.org/10.1016/j.livsci.2016.11.007
M. Yörük, M. Gül, A. Hayirli, and M. Macit, “The effects of supplementation of humate and probiotic on egg production and quality parameters during the late laying period in hens,” Poultry Science, vol. 83, no. 1, pp. 84–88, 2004. [Online]. Available: https://doi.org/10.1093/ps/83.1.84
R. Fuller, “Ecological studies on the lactobacillus flora associated with the crop epithelium of the fowl,” Journal of Applied Bacteriology, vol. 36, no. 1, pp. 131–139, 1973. [Online]. Available: https://doi.org/10.1111/j.1365-2672. 1973.tb04080.x
J. Patterson and K. Burkholder, “Application of prebiotics and probiotics in poultry production,” Poultry science, vol. 82, no. 4, pp. 627–631, 2003. [Online]. Available: https://doi.org/10.1093/ps/82.4.627
B. B. Oakley, H. S. Lillehoj, M. H. Kogut, W. K. Kim, J. J. Maurer, A. Pedroso, M. D. Lee, S. R. Collett, T. J. Johnson, and N. A. Cox, “The chicken gastrointestinal microbiome,” FEMS microbiology letters, vol. 360, no. 2, pp. 100–112, 2014. [Online]. Available: 10.1111/1574-6968.12608
P. J. Turnbaugh, V. K. Ridaura, J. J. Faith, F. E. Rey, R. Knight, and J. I. Gordon, “The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice,” Science translational medicine, vol. 1, no. 6, pp. 6ra14–6ra14, 2009. [Online]. Available: https://doi.org/10.1126/scitranslmed.3000322
E. Carbonnelle, C. Mesquita, E. Bille, N. Day, B. Dauphin, J.-L. Beretti, A. Ferroni, L. Gutmann, and X. Nassif, “Maldi-tof mass spectrometry tools for bacterial identification in clinical microbiology laboratory,” Clinical biochemistry, vol. 44, no. 1, pp. 104–109, 2011. [Online]. Available: https://doi.org/10.1016/j.clinbiochem.2010.06.017
J. M. Janda and S. L. Abbott, “16s rrna gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls,” Journal of clinical microbiology, vol. 45, no. 9, pp. 2761–2764, 2007. [Online]. Available: https://doi.org/10.1128/JCM.01228-07
U. Schillinger and F. K. Lücke, Applied and environmental microbiology, vol. 55, no. 8, pp. 1901–1906, 1989.
T. A. Tatusova and T. L. Madden, “Blast 2 sequences, a new tool for comparing protein and nucleotide sequences,” FEMS microbiology letters, vol. 174, no. 2, pp. 247–250, 1999. [Online]. Available: https://doi.org/10.1016/S0378-1097(99)00149-4
K. Sonomoto and A. Yokota, Lactic acid bacteria and bifidobacteria: current progress in advanced research. Horizon Scientific Press, 2011.
S. C. Corr, Y. Li, C. U. Riedel, P. W. O’Toole, C. Hill, and C. G. Gahan, “Bacteriocin production as a mechanism for the antiinfective activity of lactobacillus salivarius ucc118,” Proceedings of the National Academy of Sciences, vol. 104, no. 18, pp. 7617–7621, 2007. [Online]. Available: https://doi.org/10.1073/pnas.0700440104
K. A. Ryan, P. Daly, Y. Li, C. Hooton, and P. W. O’Toole, “Strain-specific inhibition of helicobacter pylori by lactobacillus salivarius and other lactobacilli,” Journal of Antimicrobial Chemotherapy, vol. 61, no. 4, pp. 831–834, 2008. [Online]. Available: https://doi.org/10.1093/jac/dkn040
C. Dunne, L. Murphy, S. Flynn, L. O?Mahony, S. O?Halloran, M. Feeney, D. Morrissey, G. Thornton, G. Fitzgerald, C. Daly et al., “Probiotics: from myth to reality. demonstration of functionality in animal models of disease and in human clinical trials,” in Lactic Acid Bacteria: Genetics, Metabolism and Applications. Springer, 1999, pp. 279–292.
S. Messaoudi, M. Manai, G. Kergourlay, H. Prévost, N. Connil, J.-M. Chobert, and X. Dousset, “Lactobacillus salivarius: bacteriocin and probiotic activity,” Food microbiology, vol. 36, no. 2, pp. 296–304, 2013. [Online]. Available: https://doi.org/10.1016/j.fm.2013.05.010
M. Velraeds, H. Van der Mei, G. Reid, and H. J. Busscher, “Inhibition of initial adhesion of uropathogenic enterococcus faecalis by biosurfactants from lactobacillus isolates.” Applied and environmental microbiology, vol. 62, no. 6, pp. 1958–1963, 1996. [Online]. Available: https://doi.org/10.1016/S0090-4295(97)00065-4
C. M. Galdeano and G. Perdigon, “The probiotic bacterium lactobacillus casei induces activation of the gut mucosal immune system through innate immunity,” Clinical and Vaccine Immunology, vol. 13, no. 2, pp. 219–226, 2006. [Online]. Available: https://doi.org/10.1128/CVI.13.2.219-226.2006
G. Storelli, A. Defaye, B. Erkosar, P. Hols, J. Royet, and F. Leulier, “Lactobacillus plantarum promotes drosophila systemic growth by modulating hormonal signals through tor-dependent nutrient sensing,” Cell metabolism, vol. 14, no. 3, pp. 403–414, 2011. [Online]. Available: https://doi.org/10.1016/j.cmet.2011.07.012
F. L. Tulini, L. K. Winkelströter, and E. C. De Martinis, “Identification and evaluation of the probiotic potential of lactobacillus paraplantarum ft259, a bacteriocinogenic strain isolated from brazilian semi-hard artisanal cheese,” Anaerobe, vol. 22, pp. 57–63, 2013. [Online]. Available: https://doi.org/10.1016/j.anaerobe.2013.06.006